Cross-species genome-wide association studies and a gene coexpression network identified genes associated with root metaxylem phenotypes in maize under water stress and non-stress and rice.
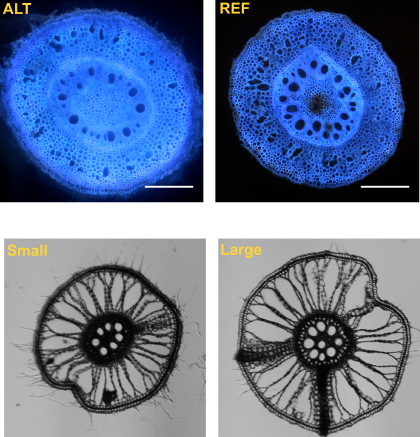
Root metaxylem phenotypes under drought stress in cereals
Authors
Klein SP, JE Reeger, SM Kaeppler, KM Brown, JP Lynch.
Source
In BioRxiv: https://www.biorxiv.org/content/10.1101/2020.11.02.365247v2
Abstract
Root metaxylem are phenotypically diverse structures whose function is related to their anatomy, particularly under drought stress. Much research has dissected the genetic machinery underlying metaxylem phenotypes in dicots, but monocots are relatively unexplored. In maize (Zea mays), a robust pipeline integrated a GWAS of root metaxylem phenes under well-watered and water stress conditions with a gene co-expression network to identify candidate genes most likely to impact metaxylem phenotypes. We identified several promising candidate genes in 14 gene co-expression modules inferred to be functionally relevant to xylem development. We also identified five gene candidates that co-localized in multiple root metaxylem phenes in both well-watered and water stress conditions. Using a rice GWAS conducted in parallel, we detected overlapping genetic architecture influencing root metaxylem phenotypes by identifying eight pairs of syntenic candidate genes significantly associated with metaxylem phenes. There is evidence that the genes of these syntenic pairs may be involved in biosynthetic processes related to the cell wall, hormone signaling, oxidative stress responses, and drought responses. Our study demonstrates a powerful new strategy for identifying promising gene candidates and suggests several gene candidates that may enhance our understanding of vascular development and responses to drought in cereals.

