RootScan is an imaging software designed to codify anatomical features of root cross sections from microscope digital images.
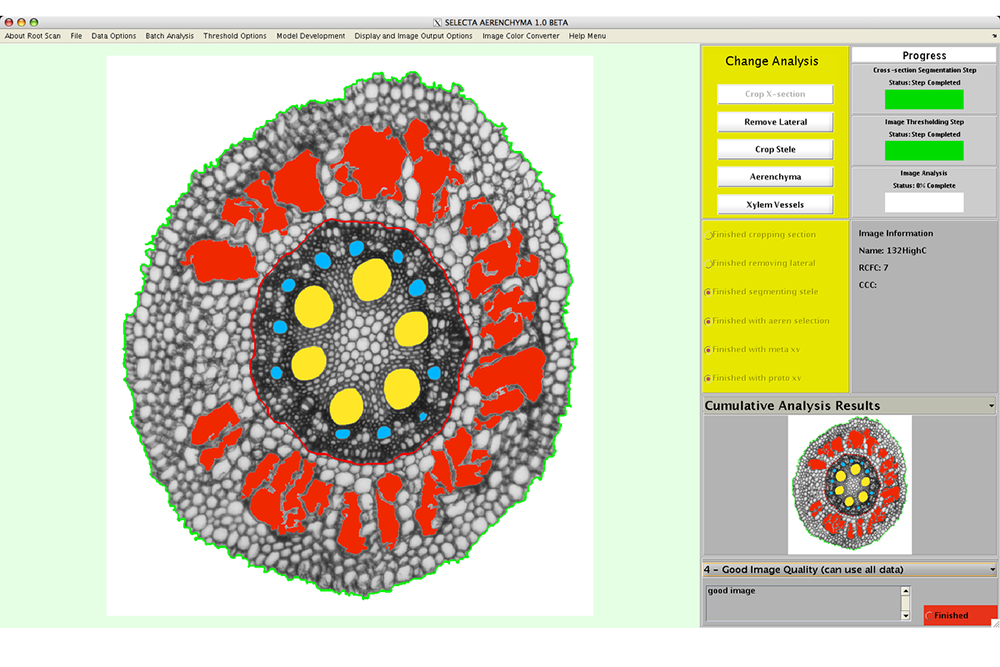
RootScan after anatomical features have been extracted and measured
RootScan is a Matlab program developed for high throughput phenotyping of anatomical traits such as aerenchyma in maize roots. The original paper describes the software and its use for root anatomical trait screening.
"RootScan2" is a Java application with updated features created by Cleoniki Kesidis for the Lynch/Brown lab in 2014, for semi-automated analysis of maize root cross section images generated via laser ablation. Improvements to this program include ability to better detect stele borders and aerenchyma, and detailed cortical cell file labeling and data output. To use RootScan2, download "RootScan2.jar" and make sure you have the latest version of Java. Source code can be made available upon request. Additional versions of RootScan2 are in progress. The following are also available for download:
RootScan2 User Manual (PDF; requires Adobe Reader)
RS2 Video Tutorial 1 : Loading files (Videos are mp4 files.)
RS2 Video Tutorial 2 : Phase 1 analysis
RS2 Video Tutorial 3 : Saving files
RS2 Video Tutorial 4 : Phase 2 analysis
RS2 Video Tutorial 5 : Phase 3 analysis
Java archive, 2.0 MB
Word 2007 document, 17.0 KB

