Discovery of root architectural and anatomical maize genes
DeepGenes research graph
Co-PIs:
Malcolm Bennett, University of Nottingham
Shawn Kaeppler, University of Wisconsin
Natalia De Leon, University of Wisconsin
Tony Bishopp, University of Nottingham
Kathleen Brown, Penn State University
Post-docs, students, staff:
Dayane Lima, Postdoc, University of Wisconsin
Vai Sa Nee Lor, Postdoc, University of Wisconsin
Riccardo Fusi, PhD student, University of Nottingham
Miranda Niemiec, PhD student, Penn State University
Jagdeep Singh Sidhu, PhD student , Penn State University
Alden Perkins, Masters student, Penn State University
Rachel Perry, Technician, University of Wisconsin
Summary
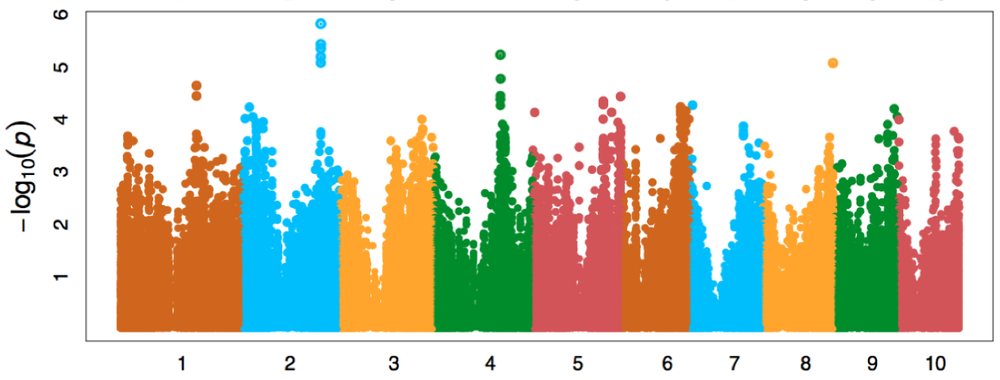
DeepGenes incorporates genes and traits related to root architecture, anatomy, and deep rooting discovered through phenotyping and LEADER into genomic selection models. Initial gene discovery was completed through GWAS of the Wisconsin Diversity panel of inbreds. Multiple genes are in various stages of validation through mutant and transgenic studies. A genomic selection model is being developed through phenotyping two panels of hybrids grown under nitrogen and water stress conditions. This model will select for the genetic structure underlying a suite of traits that we identify as being "carbon optimized."

Mutant lines target genes controlling maize root traits

