Discovery and Functional Characterization of Genes Regulating Plant Immunity in Perennial Crops (NSF 1546863)

Summary
This project will establish a new approach for use with perennial crop plants to identify candidate loci for disease resistance using a model tree crop, Theobroma cacao (the chocolate tree). Whole genome re-sequencing and transcriptome sequencing of a core collection of highly diverse cacao genotypes will provide the genetic information necessary to drive the discovery of genes critical for pathogen resistance. Functional analysis of these genes will test their role in resistance and set the stage for future translation of these basic findings to guiding more efficient breeding programs utilizing a wider array of genetic diversity. Importantly, the methods, tools, and knowledge gained will be directly applicable to discovery of genes underlying important traits in other crops, especially heterozygous perennials such as trees and many grasses which are not particularly amenable to approaches developed for the major annual crops such as corn and soybean. The basic evolutionary and functional principles that will be discovered will be  generalizable to most if not all crop plants. To promote and build genomics research capacity in developing countries and to promote inter-disciplinary cross-training the project will support scientific exchanges between project members and foreign collaborators, through postdoctoral, graduate and undergraduate student training at multiple institutions and will involve students from minority serving institutions. The results of this study will be made publicly available through electronic resources and publications.
generalizable to most if not all crop plants. To promote and build genomics research capacity in developing countries and to promote inter-disciplinary cross-training the project will support scientific exchanges between project members and foreign collaborators, through postdoctoral, graduate and undergraduate student training at multiple institutions and will involve students from minority serving institutions. The results of this study will be made publicly available through electronic resources and publications.
Background
Identifying the functional role of genetic variation in pathogen resistance is currently approached with genetic mapping and phenotyping experiments, primarily QTL, association mapping, and bulk segregant approaches. Plant species with complex genomes, difficult breeding systems, and long generation times are not well suited to these methods, and in the absence of related wild strains for introgression, there are few options available for efficient discovery of critical resistance genes and breeding for reduced susceptibility. To establish a new scientific approach to discover genetic resources for pathogen resistance in perennial crops, we propose a multi-pronged strategy to identify disease resistance genes. This approach involves integrating gene expression, evolutionary, and functional analyses to identify in an unbiased fashion the most important candidate loci for pathogen resistance. Our work will identify genes that contribute significantly to resistance in Theobroma cacao (cacao) and will serve as a model for developing and testing the feasibility of these approaches. Cacao is a tropical tree crop of significant social, ecological, and economic importance that is also under severe disease pressure. Cacao has also emerged as a leading model tropical tree species for genomics and translational biology, with a sequenced genome, many chromosomal. After validation in the proposed project, our approach may be applied to other basic biological questions in diverse species and to explore the basis of other important traits such as pest resistance and abiotic stress tolerance, and thereby contribute to meeting the challenge of increasing global food productivity.
Specific Aims
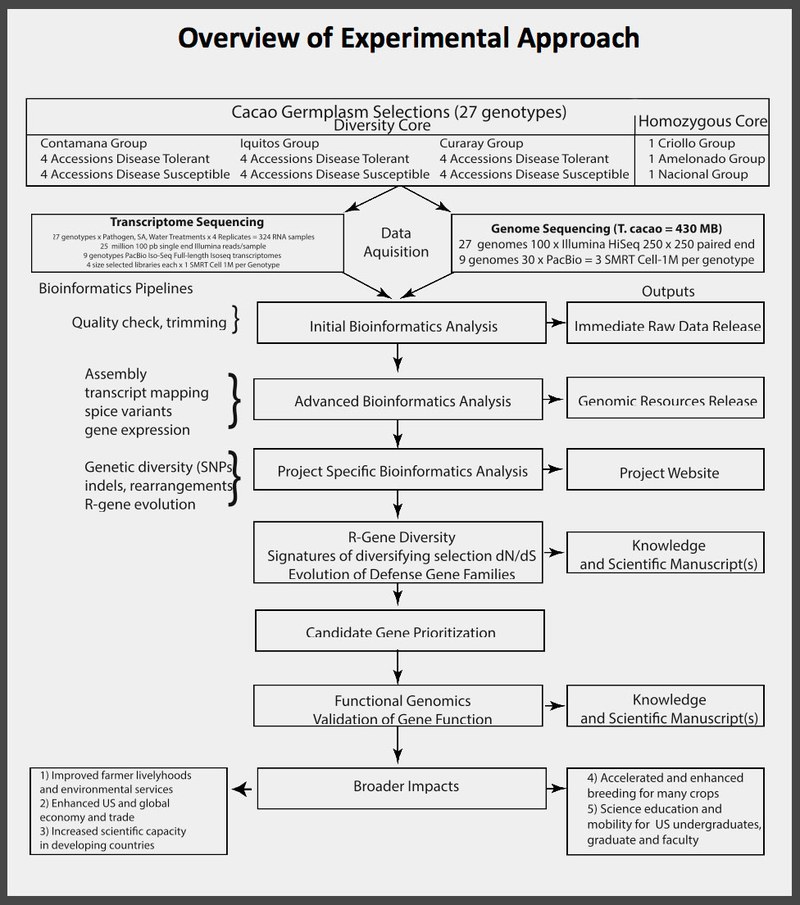
- Evaluate genomic diversity within cacao by resequencing the genomes of 8 individuals from each of 3 major clades of T. cacao. The individuals within each clade vary in resistance to Phytophthora (4 resistant, 4 susceptible). In addition, we will resequence 1 member each of three additional clades of homozygous groups of special interest, for a total of 27 cacao genomes.
- Sequence transcriptomes (RNA-Seq) to corroborate gene models, and characterize transcriptional responses to pathogen infection from the same resistant and susceptible collections of germplasm as described in 1.
- Use the newly acquired data to compare genomic and transcriptional diversity in cacao resistance-genes with gene expression and allelic diversity data from an existing collection of wild tropical trees (Marden et al., 2017). These comparisons will identify homologous (orthologous or convergent) loci possessing shared expression responses and/or signatures of adaptive evolution in multiple sympatric species. Overlap of these data will provide valuable information for identifying genes most likely to be important in affecting quantitative resistance to pathogens.
- Test the function of a set of at least 10 highly prioritized candidate resistance genes using an integrated set of transgenic approaches to validate gene function and to increase our understanding of the mechanisms regulating the plant defense response.
- Promote international collaboration, transfer of new knowledge directly to people working in cacao improvement, educate new and working scientists, and increase the impact of the NSF PGRP by involving students and scientists working together as teams from the US and developing countries in research and training programs.
Integration of Research and Education
To realize the vast potential of plant genomics in the fight against global poverty, malnutrition, starvation and environmental degradation it is essential that young scientists from the US and abroad are exposed to the opportunities for international collaboration early in their careers and are encouraged to work together on important projects. Our goals are to train and inspire a new generation of plant scientists in genomics and international research and to provide opportunities and a diverse environment for young scientists to develop collaborative networks of associates from different countries and institutions that could lead to viable long-term collaborations.
International Research Internships for Undergraduate Students: Undergraduate research develops critical thinking, creativity, problem solving and intellectual independence, promotes an innovation-oriented culture and often leads to a career in scientific research. Three undergraduate students from underrepresented groups from PSU and other institutions that serve underrepresented students across the country, and Puerto Rico will participate in 8-week summer internships during project years 2, 3 and 4 (9 internships). Undergraduate interns will form 4-person research teams with a project postdoc or a graduate student. During the internship period the students will work at PSU for the first 4 weeks to perform laboratory research, participate in PSU's specialized Spanish for the Agricultural Sciences language program, and participate in an international agricultural development orientation workshop presented by International Agriculture and Development Graduate Programs (INTAD) graduate students. Research teams will then conduct 4 weeks of international research at the Tropical Agricultural Research and Higher Education Center (CATIE) in Costa Rica.
Participants
Mark Guiltinan, PI, Penn State University
Siela Maximova, Co-PI, Penn State University
Claude dePamphillis, Co-PI, Penn State University
James Marden, Co-PI, Penn State University
Peter Tiffin, Co-PI, University of Minnesota
Dapeng Zhang, Key Collaborator, USDA ARS Beltsville
Désiré Pokou, Key Collaborator, CNRA, Ivory Coast, West Africa
Andrew Fister, Postdoctoral Scholar, Penn State University
Mariela E. Leandro Muñoz, Postdoctoral Scholar, CATIE (Centro Agronómico Tropical de Investigación y Enseñanza)
Eric Wafula, Research Bioinformatician, Penn State University
Noah Winters, Graduate Assistant, Penn State University
Lena Landherr Sheaffer, Research Technician, Penn State University
Paula Ralph, Research Technician, Penn State University
Melanie Manning, Research Technician, Penn State University
Key Publications
2018
Fister, A. S., Landherr, L., Maximova, S. N. & Guiltinan, M. J. Transient Introduction of CRISPR/Cas9 Machinery Targeting TcNPR3 Enhances Defense Response in Theobroma cacao. Plant Biotechnology Frontiers in Plant Science, March 2, 2018. https://doi.org/10.3389/fpls.2018.00268
2017
Marden JH, Mangan SA, Peterson MP, Wafula E, Fescemyer HW, Der JP, dePamphilis CW,Comita LS. 2017. Ecological genomics of tropical trees: how local population size and allelic diversity of resistance genes relate to immune responses, co-susceptibility to pathogens, and negative density dependence. Mol Ecol, 26: 2498-2513.
Immediate Raw Data Release
Links to webserver with first release datasets as per the NSF data release plan. Please read the associated Readme files for more details. This data is considered preliminary and in raw form (Immediate Raw Data Release in figure above), but has been checked for quality. Data will be periodically added to this site as it becomes available.

